Discovery and characterization of small organic molecules, cyclic peptides, and fully human antibodies specific to targets of interest.
Discovery and validation of pharmaceutical targets for ligand-based pharmacodelivery. Philochem uses state-of-the-art chemical proteomics technologies for the identification of accessible targets and for the characterization of the mechanism of action.
Development of small molecule therapeutics, with the potential to achieve an unprecedented level of activity and selectivity. Philochem is performing pioneering work for the ligand-based delivery of drugs, radionuclides and immunomodulators.
Encoded Library Technologies
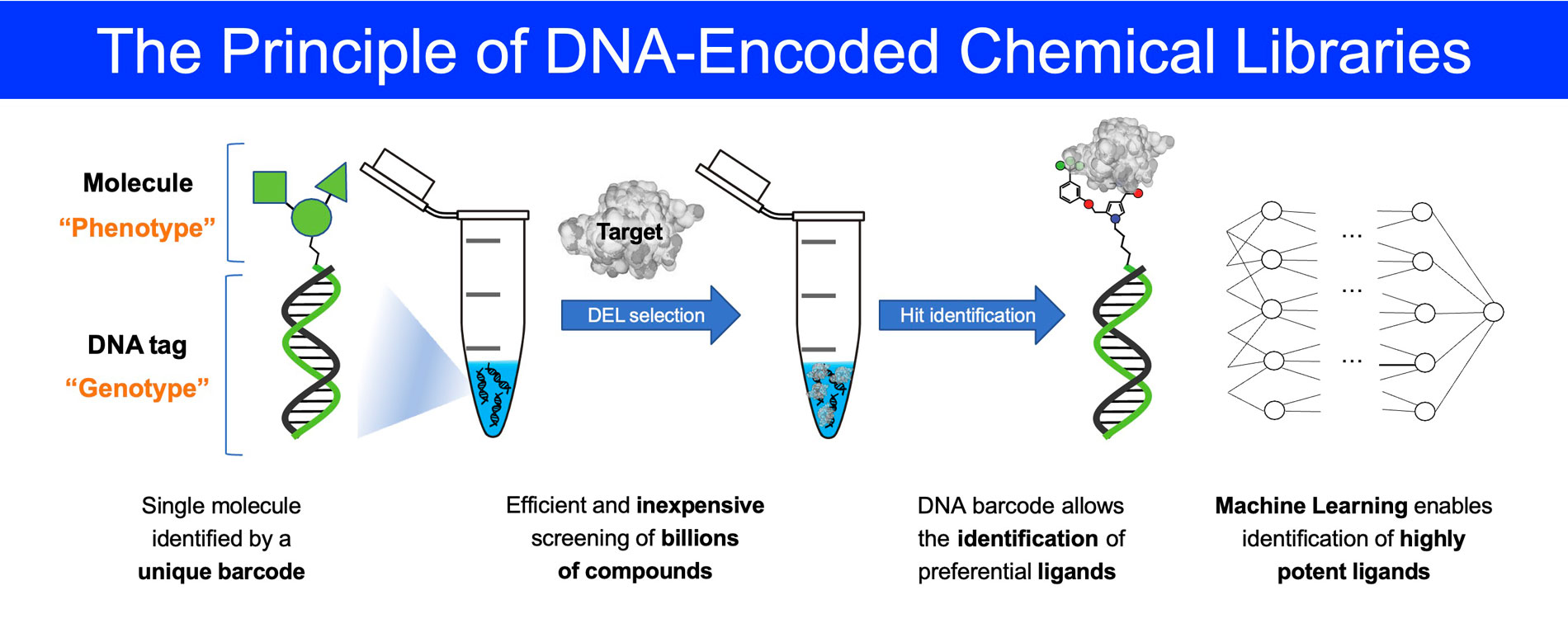
DNA-encoded chemical libraries (DELs)
The discovery of small organic ligands to protein targets has been traditionally performed by screening very large sets of organic molecules (termed “chemical libraries”), one by one, using high-throughput screening procedures. The assembly of such libraries is very expensive and time consuming. While the value of high-throughput library screening has been demonstrated in various pharmaceutical applications, it is not uncommon that binding molecules of sufficient affinity and specificity (called “hits”) cannot be discovered using conventional screening campaigns.
In light of these considerations, considerable efforts have been devoted to the establishment of encoded libraries which enable the construction and screening of compound sets of unprecedented size in a cost-efficient fashion. Encoded libraries enable a physical connection between a phenotype (i.e., a ligand) and the corresponding genetic information.
Philochem has been practicing antibody-phage display technology for the last 25 years. All antibody-ligands, that Philochem has moved to clinical trials, stem from research activities which have been conducted using its proprietary libraries.
In 2004, our group pioneered the construction of DNA-encoded combinatorial libraries of organic molecules. The technology allows the rapid selection of specific binders (“Phenotype”), physically connected to unique DNA tags (“Genotype”) that work as amplifiable identification barcodes. Since then, Philochem has synthesized several DNA-encoded chemical libraries, featuring different designs, that have yielded high affinity and selective binders to a variety of target proteins of pharmaceutical interest.
To date, Philochem uses encoded libraries for both in house discovery programs and in the frame of collaborations with pharmaceutical industrial or academic partners.
More information on Philochem´s DEL platform is available HERE
For collaboration opportunities, partnerships, or fee-for-service agreements on DNA-encoded chemical libraries, please contact Frederik.Peissert@philochem.ch
References
A DNA-encoded chemical library based on chiral 4-amino-proline enables stereospecific isozyme-selective protein recognition
Oehler et al., Nat. Chem., 2023; 15(10):1431-1443
–
Selective tumor targeting enabled by picomolar fibroblast activation protein inhibitors isolated from a DNA-encoded affinity maturation library
Puglioli et al., Chem. 2023, 9, 411-429
–
Deep Learning Approach for the Discovery of Tumor-Targeting Small Organic Ligands from DNA-Encoded Chemical Libraries
Torng et al., ACS Omega 2023, 8, 25090-25100
–
Impact of library input on the hit discovery rate in DNA-encoded chemical library selections
Puglioli et al., Chem. Sci., 2023; DOI:10.1039/D3SC03688J
–
Isolation of a Natural Killer Group 2D Small-Molecule Ligand from DNA-Encoded Chemical Libraries
Dakhel et al., ChemMedChem, 2022, 17(21):e202200350
–
Selective tumor targeting enabled by picomolar fibroblast activation protein inhibitors isolated from a DNA-encoded affinity maturation library
Puglioli et al. (2022) Chem, doi: 10.1016/j.chempr.2022.10.006
–
Dakhel et al. (2022) ChemMedChem, 17, e202200350
–
Bassi et al. (2021) J Med Chem, 64, 15799-15809
–
–
Favalli et al. (2021) Nat Chem, 13, 540-548
–
Gironda-Martinez et al. (2021) J Med Chem 64, 17496-17510
–
–
Prati et al. (2020) Biochem Biopsy’s Res Commun, 533, 235-240
–
Lerner and Neri (2020) Biochem Biopsy’s Res Commun, 527, 757-759
–
Sannino et al. (2019) Chembiochem, 20, 955-962
–
–
Dal Corso et al. (2018) Agew Chem Int Ed Engl, 57, 17178-82
–
Neri and Lerner (2018) Annu Rev Biochem, 8, 479-502
–
Li et al. (2016) ACS Comb Sci, 18, 438-43
–
Franzini et al. (2015) Anger Chem Int Ed Engl, 23, 3927-31
–
Scheuermann et al. (2015) Curr Open Chem Viol, 26, 99-103
–
Scheuermann et al. (2010) Chembiochem 11, 931-7
–
Mannocci et al. (2008) Proc Natl Acad Sci USA, 105, 17670-5
–
Encoded Library Technologies
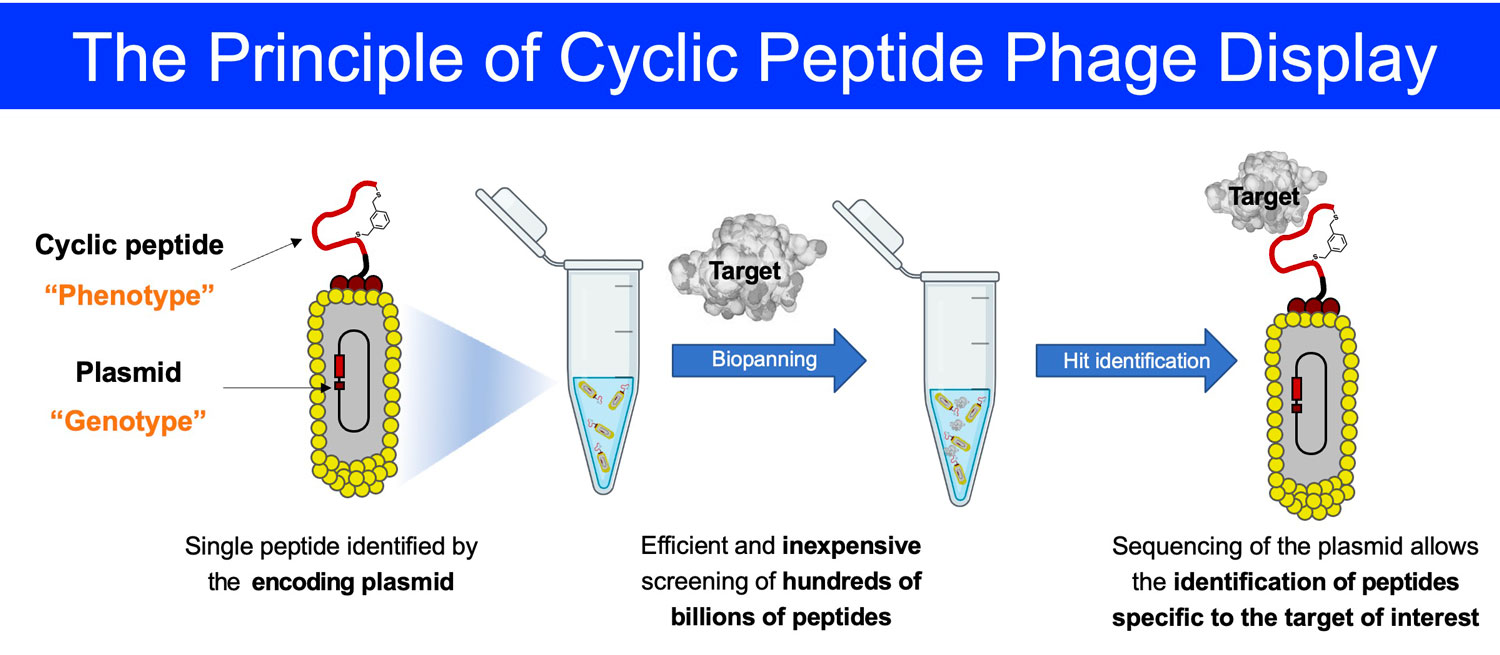
Cyclic peptide phage libraries
Cyclic peptides represent a promising avenue for the development of targeted therapeutics due to their ability to bind to target proteins with high affinity and selectivity, combined with their low inherent toxicity, their relatively high stability, and their ease of production. Cyclic peptides specific to a target of interest can be isolated from large combinatorial libraries such as phage libraries. Philochem has generated innovative cyclic peptide phage libraries comprising over 100 billion library members.
To date, Philochem uses cyclic peptide phage libraries for both in house discovery programs and in the frame of collaborations with pharmaceutical industrial or academic partners.
Encoded Library Technologies
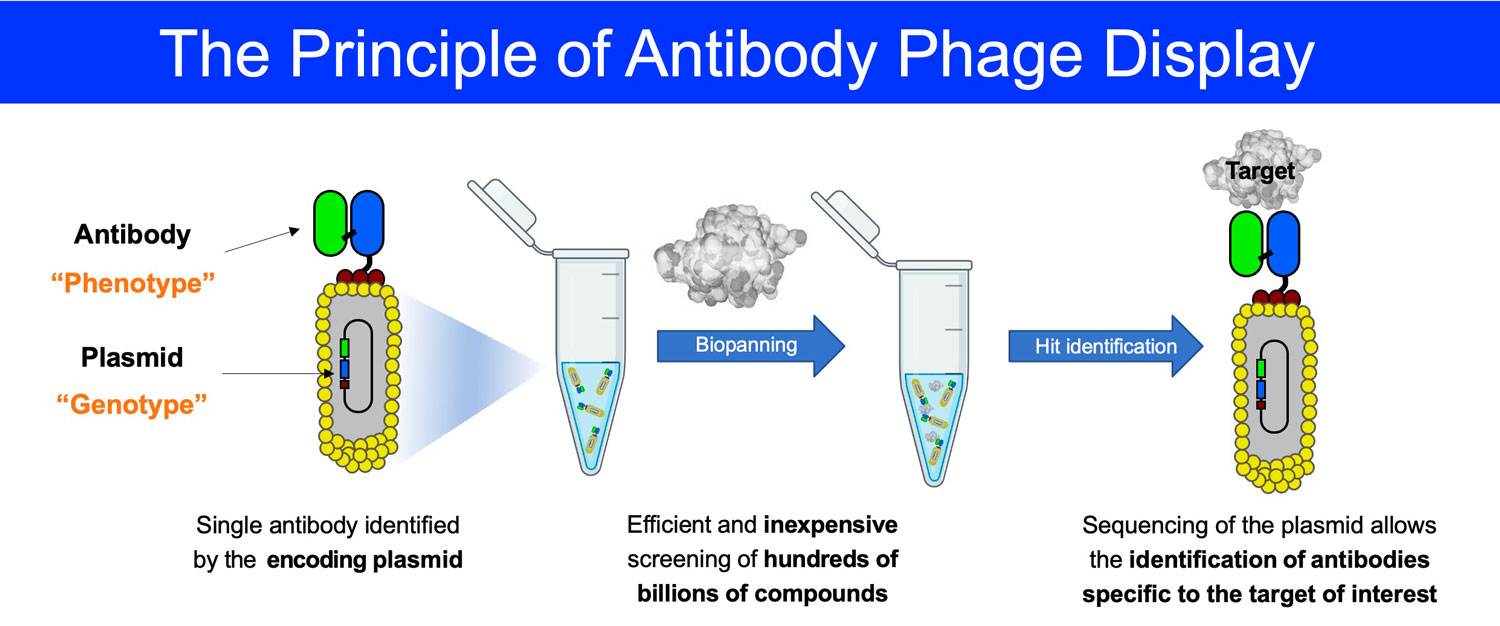
Antibody phage libraries
The discovery of specific binders to protein targets is a crucial step in the discovery of new products at Philochem. Over the last 15 years, we have published the construction and validation of numerous antibody phage libraries comprising more than 100 billion members. These proprietary libraries enable the isolation of specific human monoclonal antibodies against virtually any antigen of interest.
To date, Philochem uses antibody phage libraries for both in house discovery programs and in the frame of collaborations with pharmaceutical industrial or academic partners.
References
Generation of a novel fully human non-superagonistic anti-CD28 antibody with efficient and safe T-cell co-stimulation properties
Elsayed et al., Mabs 2023 Jan-Dec;15(1):2220839
Generation and in vivo characterization of a novel high-affinity human antibody targeting carcinoembryonic antigen
Plüss et al., Mabs 2023 Jan-Dec;15(1):2217964
Generation and in vivo validation of an IL-12 fusion protein based on a novel anti-human FAP monoclonal antibody
Nadal et al., J Immunother Cancer. 2022 Sep;10(9):e005282
Selection of a PD-1 blocking antibody from a novel fully human phage display library
Peissert et al., Protein Sci. 2022 Dec;31(12):e4486
Novel human monoclonal antibodies specific to the alternatively spliced domain D of Tenascin C efficiently target tumors in vivo
Nadal et al., MAbs. 2020 Jan-Dec;12(1):1836713
A novel synthetic naïve human antibody library allows the isolation of antibodies against a new epitope of oncofetal fibronectin
Villa et al., MAbs. 2011 May-Jun;3(3):264-72
A high-affinity human monoclonal antibody specific to the alternatively spliced EDA domain of fibronectin efficiently targets tumor neo-vasculature in vivo
Villa et al., Int J Cancer. 2008 Jun 1;122(11):2405-13
A Highly Functional Synthetic Phage Display Library Containing over 40 Billion Human Antibody Clones
Weber et al. (2014) PLoS One, 9, e100000
Selection and characterization of human antibody fragments specific for psoriasin – A cancer associated protein
Cyranka-Czaja et al. (2012) Biochem Biophys Res Commun, 419, 250-5
Design and construction of a naïve mouse antibody phage display library
Sommavilla et al. (2010) J Immunol Methods, 353, 41-3
The Extra-domain A of Fibronectin Is a Vascular Marker of Solid Tumors and Metastases
Rybak et al. (2007) Cancer Res, 67, 10948-57
Tumor-Targeting Properties of Novel Antibodies Specific to the Large Isoform of Tenascin-C
Brack et al. (2006) Clin Cancer Res, 12, 3200-8
Human monoclonal antibodies to domain C of tenascin-C selectively target solid tumors in vivo
Silacci et al. (2006) Protein Eng Des Sel, 19, 471-8
Design, construction, and characterization of a large synthetic human antibody phage display library
Silacci et al. (2005) Proteomics, 5, 2340-50
Isolation of anti-angiogenesis antibodies from a large combinatorial repertoire by colony filter screening
Giovannoni et al. (2001) Nucleic Acids Res, 29, E27
Design and use of phage display libraries for the selection of antibodies and enzymes
Viti et al. (2000) Methods Enzymol, 326, 480-505
A High-Affinity Human Antibody That Targets Tumoral Blood Vessels
Tarli et al. (1999) Blood, 94, 192-8
Human Antibodies With Subnanomolar Affinity Against a Marker of Angiogenesis Eluted from a Two-Dimensional Gel
Pini et al. (1998) J Biol Chem, 273, 21769-76
Targeting by affinity–matured recombinant antibody fragments of an angiogenesis associated fibronectin isoform
Neri et al. (1997) Nat Biotechnol, 15, 1271-5
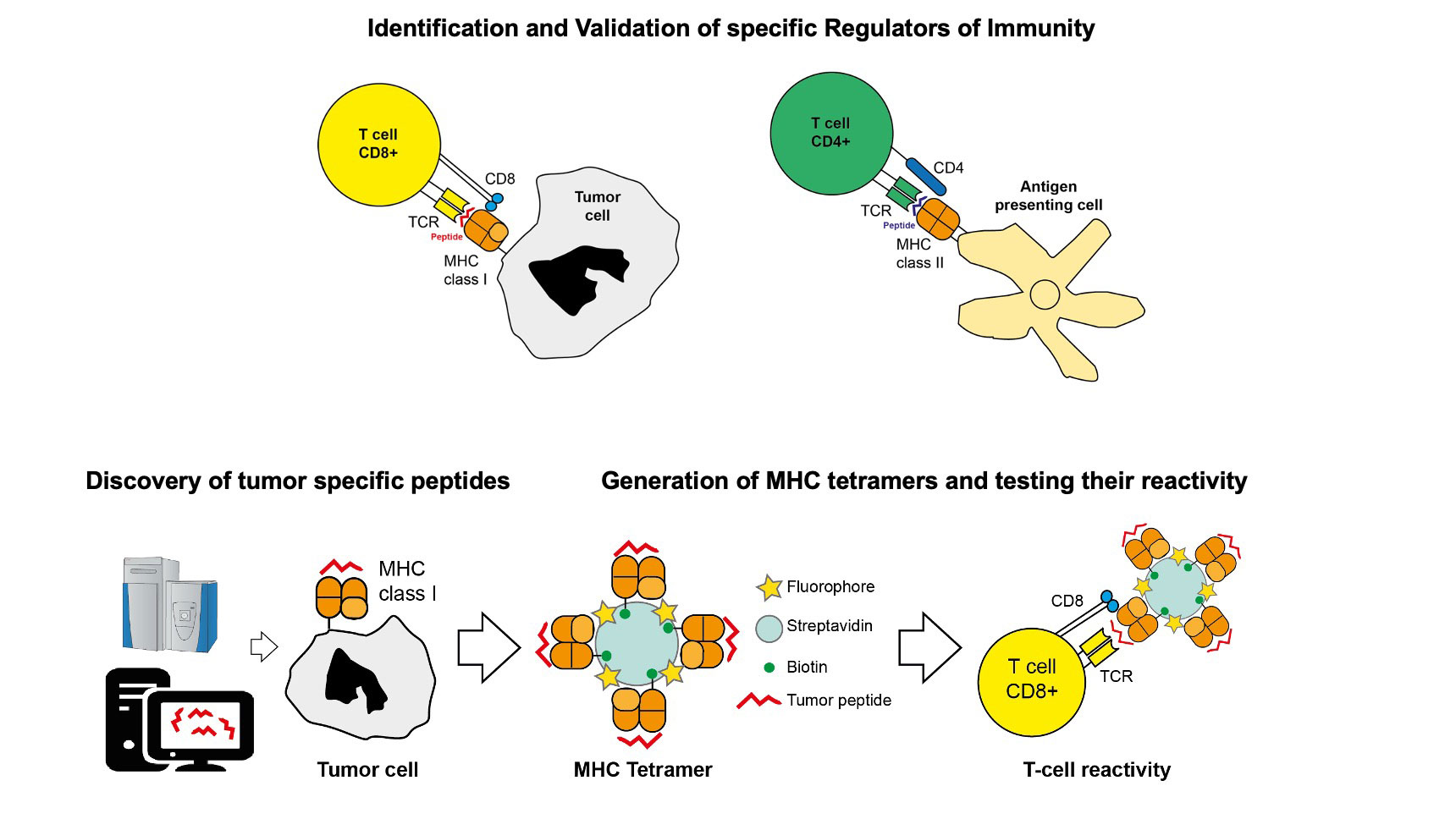
2. Discovery and validation of pharmaceutical targets
The development of targeted therapeutics crucially relies on the identification of target antigens, which are specifically expressed at the site of disease, and which are readily accessible for agents coming from the bloodstream.
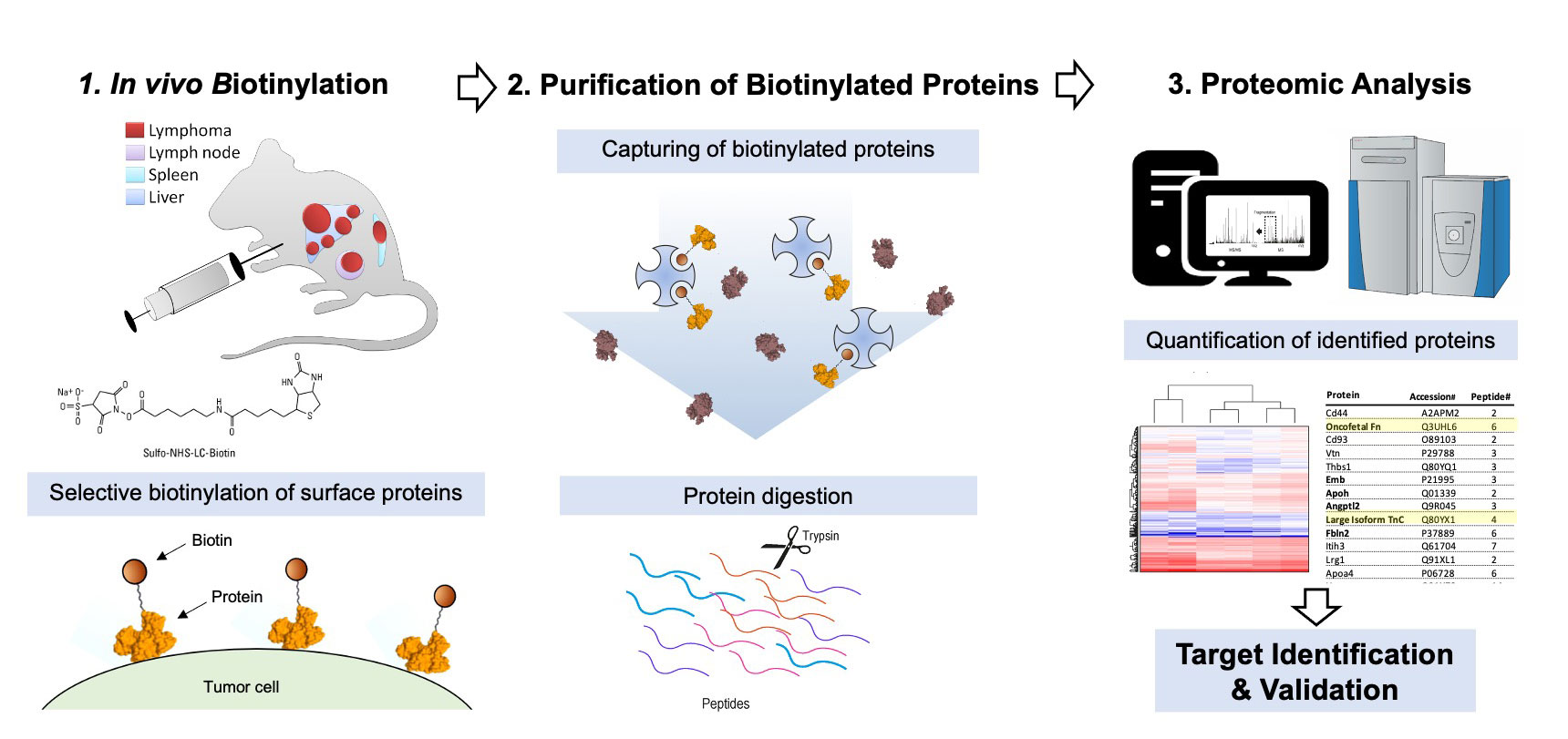
In collaboration with with ETH Zürich, Philochem has pioneered chemical proteomics methods for the characterization of accessible markers of pathology, which can be drugged by antibodies and small molecule ligands.
In a simple setup, cell lines can be submitted to surface biotinylation followed by capture on streptavidin resin and mass spectrometric analysis of tryptic peptides.
In a more complex setup, in vivo biotinylation procedures of rodent models of pathology enable the chemical proteomic characterization of vascular proteins in health and in disease. The technology has been extended to the ex vivo perfusion of surgically-resected human organs with cancer.
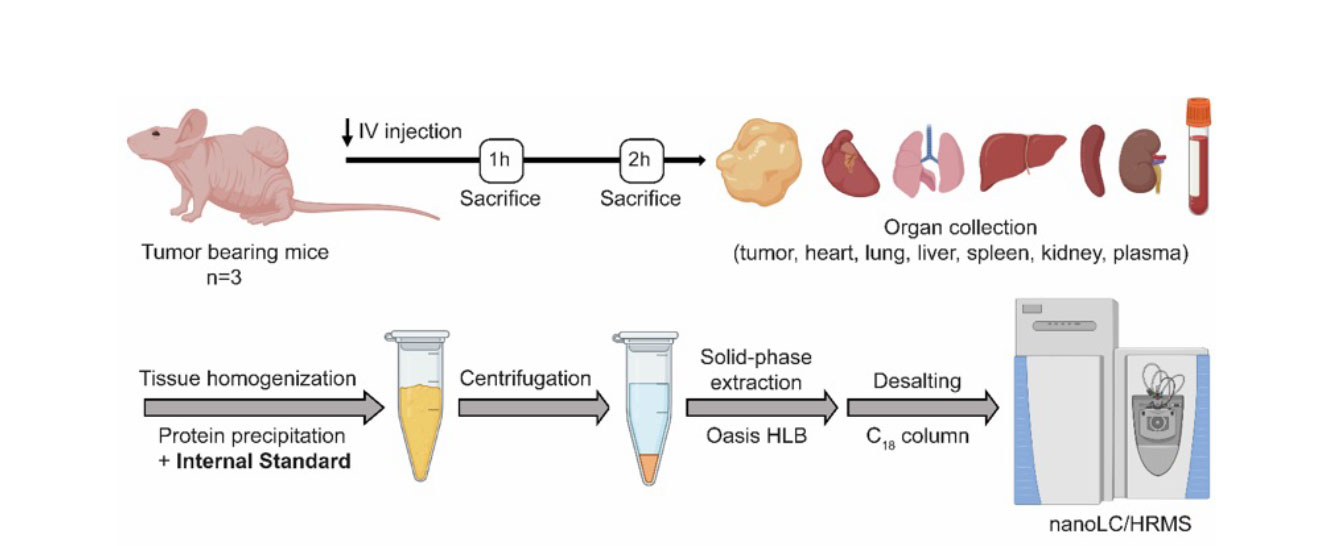
More broadly, we use MS-based chemical proteomics approaches to study the mechanism of action of our drugs. This includes the mass spectrometric quantification of drug delivery and release, as well as MHC-I peptidome analysis
References
Mass Spectrometry-Based Method for the Determination of the Biodistribution of Tumor-Targeting Small Molecule–Metal Conjugates
Gilardoni et al. (2022) Anal Chem, 93, 10715
–
Antibody-based Delivery of TNF to the Tumor Neovasculature Potentiates the Therapeutic Activity of a Peptide Anticancer Vaccine
Probst et al. (2019) Clin Cancer Res, 25, 698-709
–
Probst et al. (2017) Cancer Res, 77, 3644-54
–
Ritz et al. (2017) Proteomics, 17, 10.1002/pmic.201600364
–
Gloger et al. (2016) Cancer Immunol Immunother, 65, 1377-93
–
Ritz et al. (2016) Proteomics, 16, 1570-80
–
Sofron et al. (2016) Eur J Immunol, 46, 319-28
–
Elia et al. (2014) J Proteomics, 107, 50-5
–
Neri and Supuran (2011) Nat Rev Drug Discov, 10, 767-77
–
Fugmann et al. (2011) Kidney Int, 80, 272-81
–
–
Schliemann et al. (2010) Blood, 115, 736-44
–
Borgia et al. (2010) Cancer Res, 70, 309-18
–
–
Rösli et al. (2008) Methods Mol Biol, 418, 89-100
–
Conrotto et al. (2008) Int J Cancer, 123, 2856-64
–
Castronovo et al. (2006) Mol Cell Proteomics, 5, 2083-91
–
Brack et al. (2006) Clin Cancer Res, 12, 3200-8
–
Scheurer et al. (2005) Proteomics, 5,2718-28
–
Rybak et al. (2005) Nat Methods, 2, 291-98
–
Neri and Bicknell (2005) Nat Rev Cancer, 5,436-46
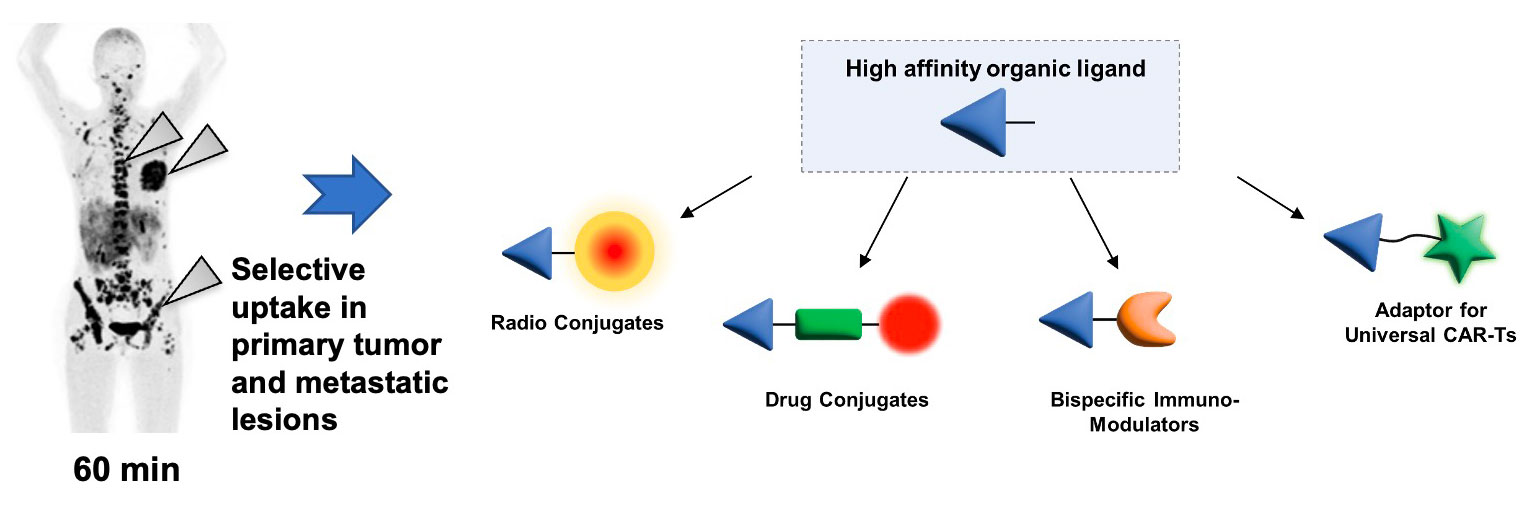
Small Molecule Therapeutics
Low molecular weight compounds represent attractive alternatives to antibodies for tumor targeting applications. The advantages of small molecule therapeutics include a fast extravasation, a deep and homogeneous penetration into solid tumors, as well as faster development timelines.
Thanks to technological advances in the field of DNA-encoded chemical libraries, we are able to generate small molecule ligands with antibody-like properties featuring ultra-high affinity to their target antigen and long residence time at the tumor site.
Philochem uses small ligands with exceptional targeting performance for the selective delivery of (i) radionuclides, (ii) cytotoxic agents, (ii) immunomodulators, or (iv) adaptors for universal Chimeric Antigen Receptor T cell therapy.
Please visit our pipeline section for more information about individual programs.
References
Puglioli et al. (2022) Chem, doi: 10.1016/j.chempr.2022.10.006
–
Zana et al. (2022), Clin Cancer Res CCR-22-1788
–
Lucaroni et al. (2022) Eur J Nucl Med Mol Imaging, doi: 10.1007/s00259-022-05982-8
–
Galbiati et al. (2022) J Nucl Med, numed.122.264036
–
Backhaus et al. (2021) Eur J Nucl Med Mol Imaging, 10.21203/rs.3.rs-969176/v1
–
Millul et al. (2021) PNAS, 118, 16, e2101852118
–
Millul et al. (2021) Mol Cancer Ther, 20, 512-22
–
Kulterer et al. (2021) J Nucl Med, 62, 360-5
–
Pellegrino et al. (2020) Bioconjug Chem, 31, 1775-83
–
Cazzamalli et al. (2018) Clin Cancer Res, 24, 3656-67
–
Cazzamalli et al. (2017) J Control Release, 246, 39-45
–
Cazzamalli et al. (2016) Mol Cancer Ther, 15, 2926-35
–
Krall et al. (2016) J Nucl Med, 57, 943-9
–
Wichert et al. (2015) Nat Chem, 7, 241-9
Casi & Neri (2015) J Med Chem, 58, 8751-61
–
Minn et al. (2016) Oncotarget, 7, 56471-79
–
Yang et al. (2015) Oncotarget, 6, 33733-42
–
Krall et al. (2014) Chem Sci, 5, 3640-4
–
Krall et al. (2014) Angew Chem Int Ed, 53, 4231-5
–
Krall et al. (2013) Angew Chem Int Ed Engl, 52, 1384-1402
–
Müller et al. (2013) J Nucl Med, 54, 124-31
–
Ahlskog et al. (2009) Bioorg Med Chem Lett, 19, 4581-6
–
Dumelin et al. (2008) Angew Chem Int Ed Engl, 47, 3196-3201
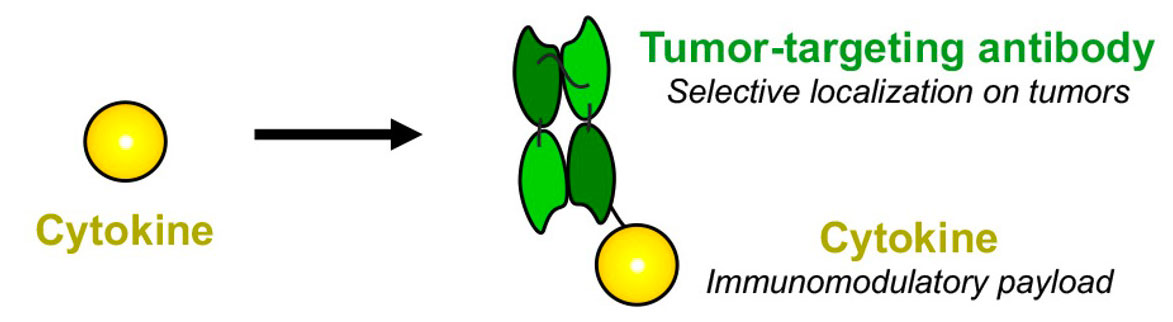
Antibody-based Therapeutics
The Philogen Group has pioneered fully human antibody-cytokine fusion proteins (also called “immunocytokines”). Cytokines are naturally produced proteins of the human body, which can either potently boost or suppress the activity of the immune system. A number of recombinant cytokines have been approved for therapeutic use, but their systemic administration at therapeutically active doses is often hindered by strong side effects. To overcome this limitation, we have engineered innovative antibody-cytokine fusion proteins to selectively localize at the site of disease and to obtain an improved therapeutic index.
The cytokine payload is chosen depending on the nature of the disease. For example, pro-inflammatory cytokines such as TNF, IL2 or IL12 can be used in oncology, while anti-inflammatory cytokines such as IL10 can be used to treat inflammatory conditions.
Our Group has generated, produced, tested, and published more than 50 cytokine payloads fused to proprietary targeting antibodies. The most promising candidates have been brought to clinical trials, one of which has recently successfully completed a Phase III trial.
For more information about the individual programs, please visit the pipeline section of the Philogen group.
References
Look et al. (2023) Sci Transl Med 15(697):eadf2281
Plüss et al. (2023), Mabs 15(1):2217964
Di Nitto et al. (2023), Pharmaceutics 15(2):377
Heiss et al. (2023), Eur J Clin Invest 53(3):e13907
Prodi et al. (2023) Antibodies (Basel), 12(2):29
Nadal et al. (2022) J Immunother Cancer 10(9):e005282
Di Nitto et al (2022) Antibodies (Basel) 11(1):19
Pimentel et al. (2021) J Immunother Cancer, 9(3):e001764.
Ongaro et al (2021) J Struct Biol 213(1):107696
Gouyou et al. (2021) Exp Biol Med 246(8):940-951
Gouyou et al. (2021) Int J Mol Sci 22(7):3460
Stringhini et al. (2021) Mabs 13(1):1868066
Corbellari et al. (2021) Mol Cancer Ther, 20, 859-71
Ongaro et al. (2020) Oncotarget, 11, 3698-3711
Weiss et al. (2020) Sci Transl Med, 7, 12(564):eabb2311
Neri (2019) Cancer Immunol Res, 7, 348-54
Hutmacher and Neri (2019) Adv Drug Deliv Rev, 15, 67-91
De Luca et al. (2017) Mol Cancer Ther, 16, 2442-51
Bootz and Neri (2016) Drug Discov Today, 21, 180-9
Kiefer and Neri (2016) Immunol Rev, 270, 178-92
Neri and Sondel (2016) Curr Opin Immunol, 40, 96-102
Hemmerle and Neri (2014) Int J Cancer, 134, 467-77
Hemmerle et al. (2014) Proc Natl Acad Sci USA, 111, 12008-12
Gutbrodt et al. (2013) Sci Transl Med, 5, 201ra118
Schwager et al. (2013) J Invest Dematol, 133, 751-8
Moschetta et al. (2012) Cancer Res, 72, 1814-24
Pasche and Neri (2012) Drug Discov Today, 17, 583-90
Pasche et al. (2012) Clin Cancer Res, 18, 4092-103;
Pasche et al. (2012) Clin Cancer Res, 18, 4092-103;
Marlind et al. (2008) Clin Cancer Res, 14, 6515-24